Research interests

Single protein-DNA complexes
Living cells carry thousands of nanomachines that assemble, disassemble, transport or process biomolecules. We study machines of gene expression (the path from genes on DNA to functional proteins) by observing single molecules of gene-expression machinery. Our main tool is single-molecule fluorescence spectroscopy, a technique that can measure nanometre distances and study molecular interactions in real time (as "molecular movies"). We also study artificial machines and devices, such as DNA-based nanodevices. Our work is multidisciplinary, combining optics, spectroscopy, biochemistry, molecular biology, molecular modelling and signal processing.
1. Mechanisms of gene transcription
Gene Machine: bacterial RNA polymerase
1a. In vitro transcription studies
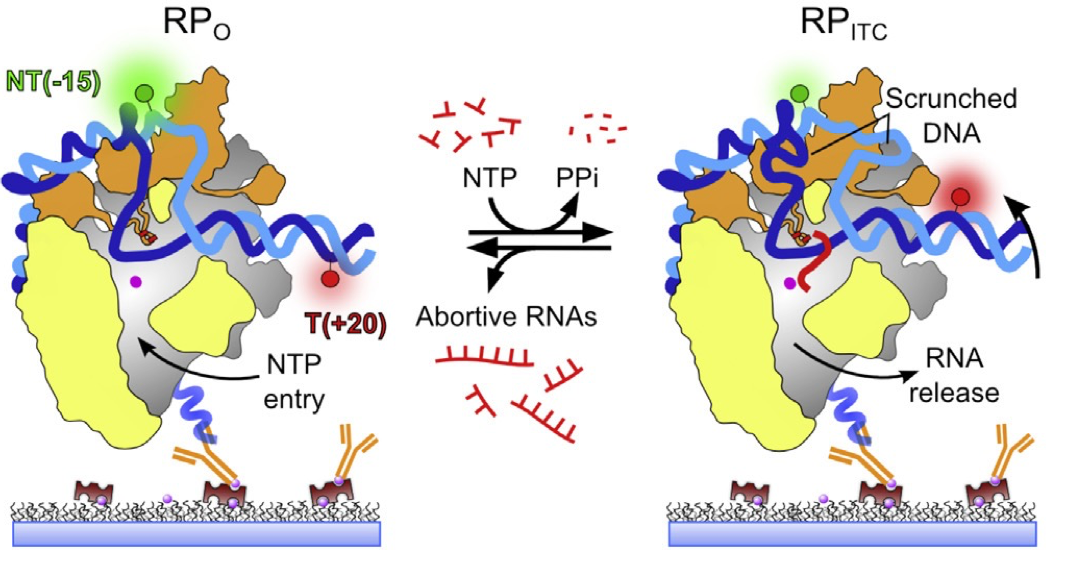
RNAP-DNA open complex
Transcription is the first and most important step in gene expression, the information flow from DNA to RNA to proteins. Most of the regulation of gene expression occurs at the level of transcription, which makes this process vital for dissecting basic genetic mechanisms, for understanding diseases due to de-regulation of gene expression, and for building synthetic gene networks and organisms for a variety of applications. Read more...
1b. In vivo studies: RNA polymerase and other DNA-binding proteins
An Escherichia coli cell with four RNA polymerase tracks.
Transcription and replication are two of the most fundamental processes necessary for life. In order to really understand how these complex mechanisms work, we need to be able to image processes taking place inside individual living cells. Fluorescence microscopy is a powerful method for doing this; however, like all light microscopy, it suffers from a fundamental limit to the achievable resolution, known as the diffraction limit. For visible light, this is around 200 nm. The processes involved in transcription and replication happen on a scale below this threshold and remains out of reach of traditional microscopy, but we can beat this limit with new 'super-resolution' techniques. Read more...
2. Mechanisms of influenza virus replication and transcription
Gene Machine: Influenza RNA polymerase
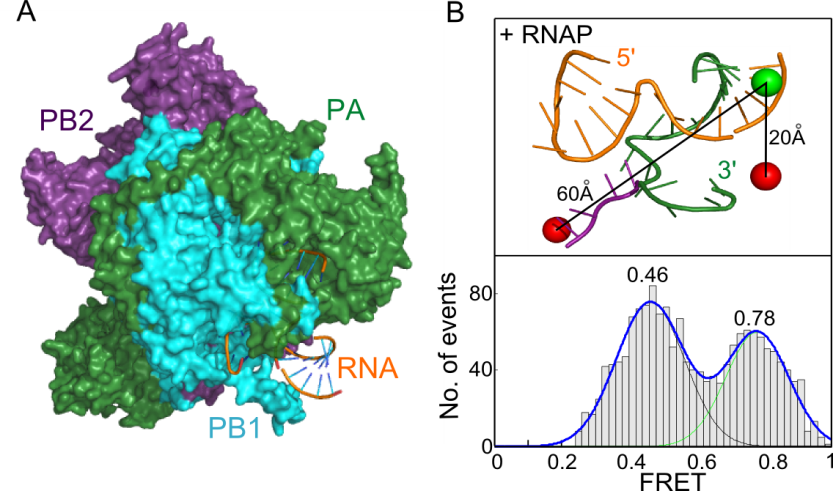
Influenza is one of the world’s major uncontrolled diseases, the cause in humans of both mild annual epidemics and more severe intermittent pandemics, such as the ‘swine flu’ outbreak in 2009 and potentially H5N1 ‘bird flu’. Influenza is caused by the influenza virus, a member of the Orthomyxovirus family. In spite of their importance, many aspects of influenza virus replication are poorly understood. Read more...
3. Mechanisms of DNA repair and synthesis
Gene Machine: DNA polymerase
DNA Polymerase I
DNA polymerases are dynamic molecular machines that faithfully copy DNA during DNA replication and repair.[1] One of the important properties of this enzyme is its remarkable fidelity, which is due to a series of non-covalent transitions that precede the chemical step of phosphoryl transfer and serve as kinetic checkpoints, rejecting inappropriate substrates early in the reaction pathway[2]. An important conformational change is the “fingers-closing” transition, inferred from cocrystal structures, in which the addition of the correct complementary deoxyribonucleotide (dNTP) to a polymerase-DNA (Pol-DNA) binary complex results in a transition from an open to a closed conformation, forming a snug binding pocket around the nascent base pair. The small magnitude of such conformational changes, coupled with the transient nature of the intermediates, makes ensemble study the fingers-closing transitions especially difficult. Read more...
4. Mechanisms of DNA recombination
Gene Machine: DNA recombinases
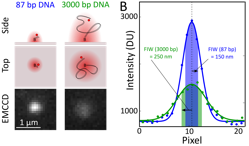
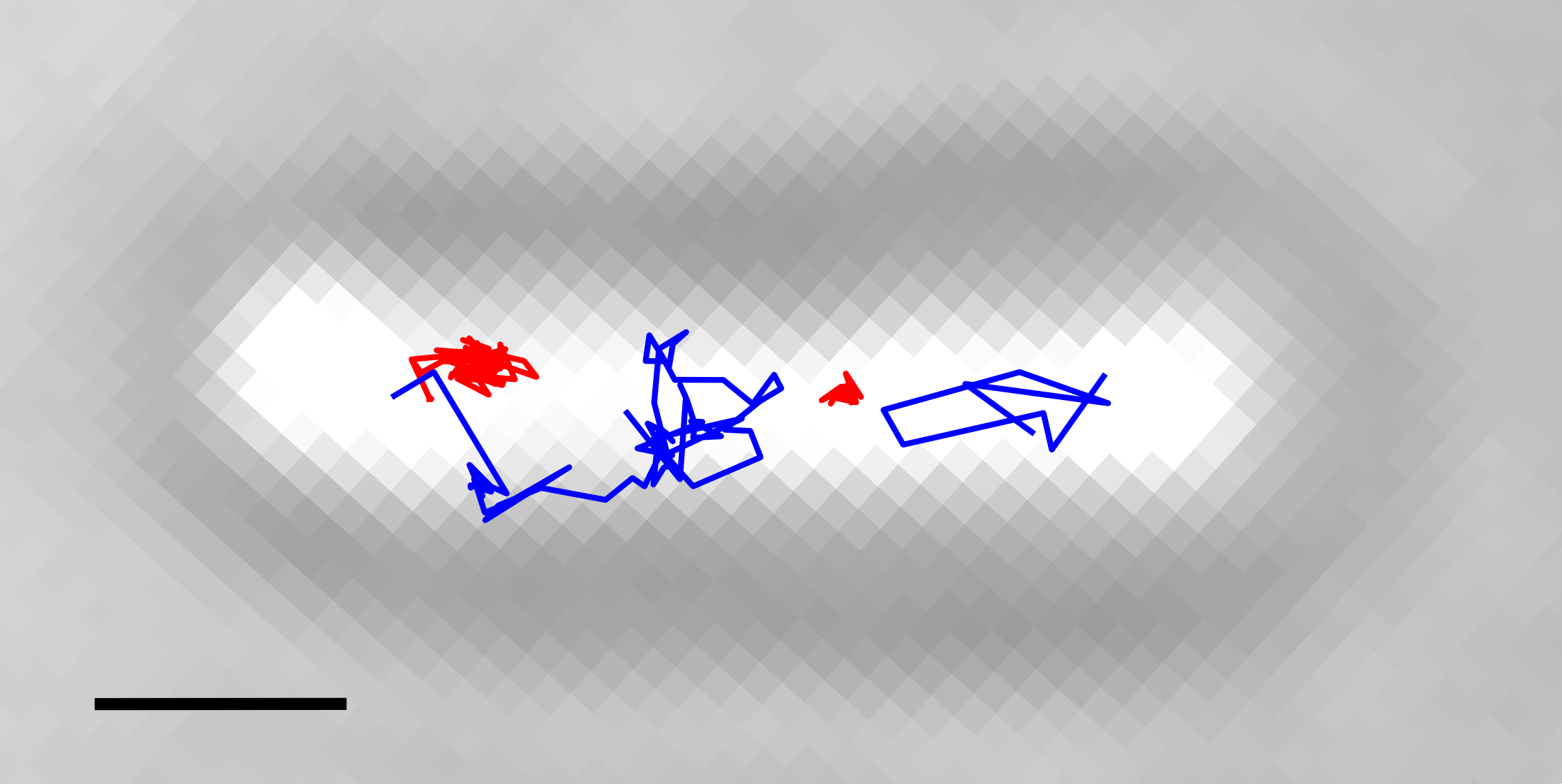
Observables of tethered-fluorophore motion for a DNA sample.
Site-specific DNA recombination is the protein-mediated breakage, exchange and rejoining of two double stranded DNA molecules at specific sequences. Site-specific recombinases carry out a wide variety of biologically important functions including: integration and excision of viral genomes, resolutions of bacterial chromosome dimers, and the regulation the expression of some genes (1). Two families of site-specific recombinases exist, named after the amino acid residue that cleaves the DNA backbone: tyrosine and serine. Read more...
5. Biosensors for DNA, RNA and pathogen detection
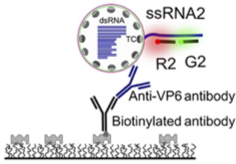
A single-molecule FRET-based strategy to detect specific viral RNAs.
DNA is an extremely versatile material with which to build biosensors; molecules can be encoded with binding sites specific for proteins of interest, or for other nucleic acids (DNA/RNA), and can be easily modified to contain useful chemical groups such as fluorophores, biotin or quantum dots. Our lab have been developing biosensors for the detection of DNA-binding proteins (1-2), nucleic acids (3) and pathogens, such as viruses (3) and bacteria. Read more...
6. Methods: single-molecule imaging and super-resolution microscopy
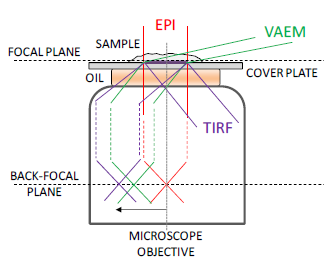
Near surface illumination techniques
In our lab, we use a variety of single molecule techniques to study biomolecular interactions. Single-molecule fluorescence spectroscopy (SMFS) allows us to study the interactions of relevant biological species with the use of Förster resonance energy transfer (FRET) combined with alternating laser excitation (ALEX). Read more...
Other projects in gene expression are under development. Please contact Dr. Kapanidis for project descriptions.








