1b. In vivo studies: RNA polymerase and other DNA-binding proteins
Transcription and replication are two of the most fundamental processes necessary for life. In order to really understand how these complex mechanisms work, we need to be able to image processes taking place inside individual living cells. Fluorescence microscopy is a powerful method for doing this; however, like all light microscopy, it suffers from a fundamental limit to the achievable resolution, known as the diffraction limit. For visible light, this is around 200 nm. The processes involved in transcription and replication happen on a scale below this threshold and remains out of reach of traditional microscopy, but we can beat this limit with new 'super-resolution' techniques.
We use a technique called Photoactivated Localisation Microscopy (PALM). This works by using cells in which molecules of interest have been labelled with photoactivatable fluorescent proteins (1). Instead of imaging all the fluorescent proteins in a cell at once, only a very small subset are activated at any given time. This allows us to localise the image produced by individual fluorescent proteins using fitting algorithms. By combining the centroid position of each fit, we can create an overall image with a much greater resolution than traditional microscopes.
The microscope we use can locate individual FPs with an accuracy of around 40 nm, providing an order of magnitude improvement on traditional microscopy. But our methods don't just give more accurate positions of molecule within a cell; we can also track the movement of individual molecules, which gives us information not just on their location but also their copy numbers, their diffusive motion and even on their enzymatic activity.
Using these techniques we are investigating the distribution of nucleic acid polymerases in Escherichia coli cells (2-6). In one such example (5), we used PALM to study the localization and dynamics of the transcription machinery and DNA in live bacterial cells, at both the single-molecule and the population level. We used photoactivated single-molecule tracking to discriminate between mobile RNAPs and RNAPs specifically bound to DNA, either on promoters or transcribed genes. We found that mobile RNAPs can explore the whole nucleoid while searching for promoters, and spend 85% of their search time in nonspecific interactions with DNA. On the other hand, the distribution of specifically bound RNAPs shows that low levels of transcription can occur throughout the nucleoid. Further, clustering analysis and 3D structured illumination microscopy (SIM) showed that dense clusters of transcribing RNAPs form almost exclusively at the nucleoid periphery. Our findings also showed that transcription can cause spatial reorganization of the nucleoid, with movement of gene loci out of the bulk of DNA as levels of transcription increase. This work provided a global view of the organization of RNA polymerase and transcription in living cells, and paved the way for more detailed analysis of the observed RNAP clusters and their functional properties.
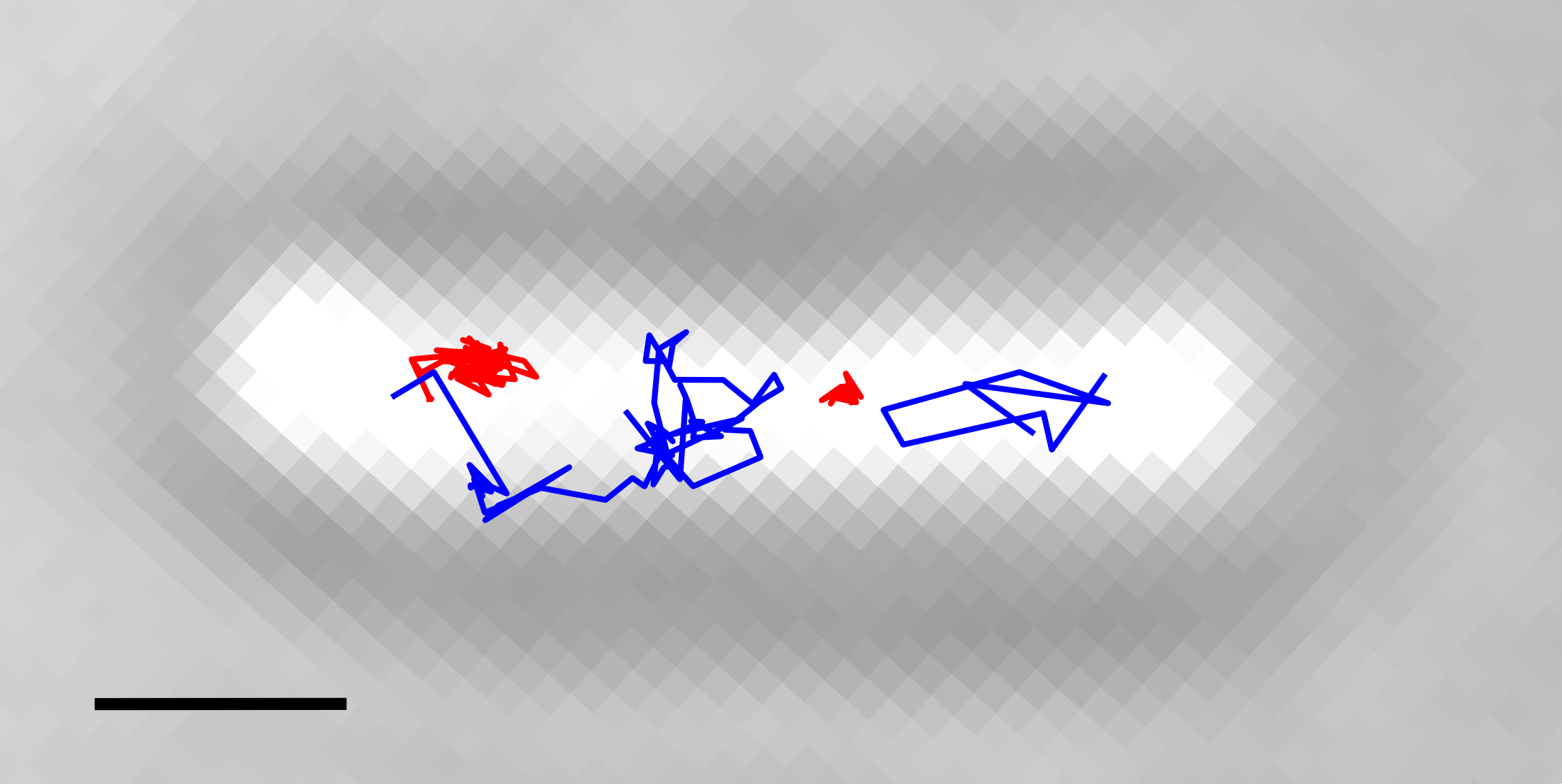
An Escherichia coli cell with four examples of RNA polymerase tracks. Tracks in red show immobile molecules and tracks in blue show mobile molecules. Scale bar, 1 μm.
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006, 313(5793), 1642–5.
- Stracy M, Uphoff S, Garza de Leon F, and Kapanidis AN. In vivo single-molecule imaging of bacterial DNA replication, transcription, and repair. FEBS Letters. 2014, 588(19), 3585–3594.
- Stracy M, Lesterlin C, Garza de Leon F, Uphoff S, Zawadzki P, and Kapanidis AN. Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. PNAS. 2015, 112(32), 201507592.
- Zawadzki P, Stracy M, Ginda K, Zawadzki K, Lesterlin C, Kapanidis AN and Sherratt D J. The Localization and Action of Topoisomerase IV in Escherichia coli Chromosome Segregation Is Coordinated by the SMC Complex, MukBEF. Cell Reports. 2015, 13(11), 2587–2596.
- Stracy M, Jaciuk M, Uphoff S, Kapanidis AN, Nowotny M, Sherratt DJ and Zawadzki, P. Single-molecule imaging of UvrA and UvrB recruitment to DNA lesions in living Escherichia coli. Nature Communications. 2016, 7, 12568.
- Endesfelder U, Finan K, Holden SJ, Cook PR, Kapanidis AN, Heilemann M, Multiscale Spatial Organization of RNA Polymerase in Escherichia coli., Biophys. J., 2013; 52 (8), pp.172-181.








