Mechanisms of site-specific DNA recombination
Justin Pinkney, Pawel Zawadzki, David Sherratt and Achillefs Kapanidis
Site-specific recombination is the protein mediated breakage, exchange and rejoining of two double stranded DNA molecules at specific sequences. Site-specific recombinases carry out a wide variety of biologically important functions including: integration and excision of viral genomes, resolutions of bacterial chromosome dimers, and the regulation the expression of some genes (1). Two families of site-specific recombinases exist, named after the amino acid residue that cleaves the DNA backbone: tyrosine and serine.
Tyrosine recombinases In collaboration with David Sherratt’s group in Oxford biochemistry we are using novel single-molecule fluorescence techniques to study two site-specific recombination proteins: Cre and XerCD. These proteins sequentially exchange pairs of DNA strands to form a Holliday junction intermediate followed by resolution of the Holliday junction to form the recombinant product DNA. We have developed a novel method to use single-molecule fluorescence to observe large scale conformational changes of DNA called Tethered Fluorophore Motion (TFM). Combining this technique with single-molecule FRET has allowed us to study tyrosine recombinases in unprecedented detail, observing both the large and small scale conformational changes as recombination occurs in real time at the slide surface.
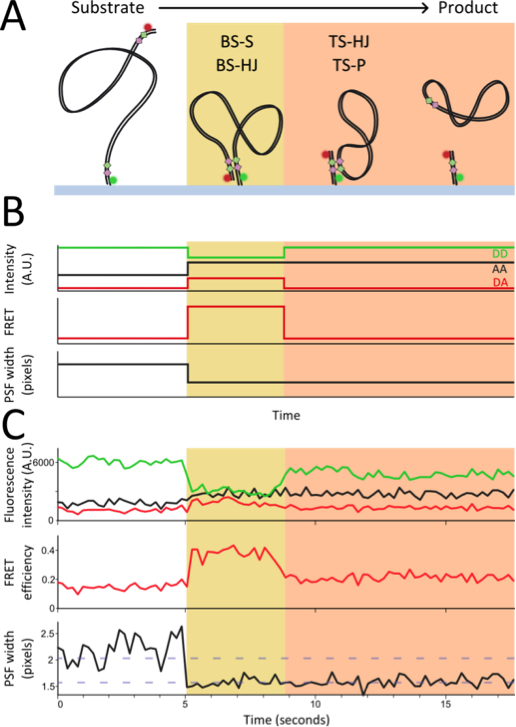
(A) Schematic of Cre recombination on surface immobilised DNAs. (B) Expected FRET and TFM observables during each stage of the reaction. (C) Representative single-molecule time trace of Cre recombination.
Serine recombinases Serine recombinases operate in a strikingly different manner to the tyrosine recombinases described above. All four DNA strands are cut simultaneously and the intriguing mechanism for strand exchange is thought to involve the 180° rotation of one half of the complex, held together by hydrophobic protein-protein interactions (3). In collaboration with Prof. Nigel D.F. Grindley at Yale University we hope to use single-molecule FRET to study two serine recombinase proteins: gamma-delta resolvase and Bxb1 integrase. We will use single-molecule FRET measurements to identify the intermediates formed during recombination and to study the dynamics and structural changes involved in the strand exchange process.
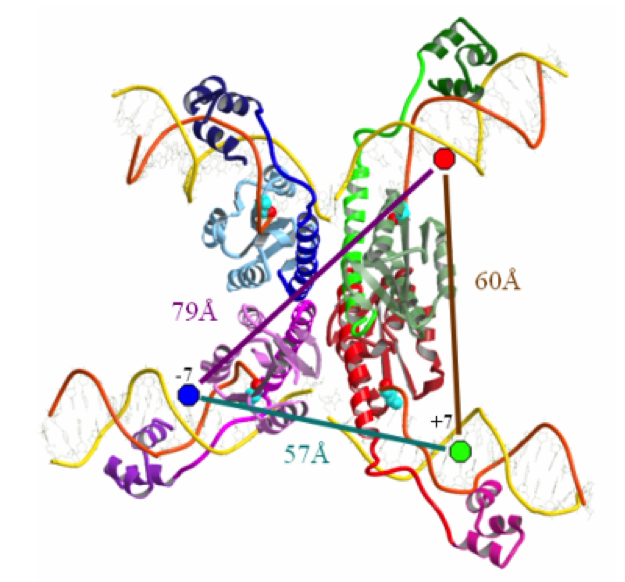
Structure of the gamma-delta resolvase synaptic complex. View through the complex showing the flat interface that separates the complex into two halves. Blue, green and red circles represent fluorophore labels on the DNA strands and their distances between them, as measured from the crystal structure.
- Grindley NDF, Whiteson KL, Rice PA (2006) Mechanisms of site-specific recombination. Annual Review of Biochemistry 75:567–605.
- Pinkney JNM et al. (2012) Capturing reaction paths and intermediates in Cre-loxP recombination using single-molecule fluorescence. Proceedings of the National Academy of Sciences of the United States of America:1211922109–.
- Bai H et al. (2011) Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase. Proceedings of the National Academy of Sciences 108:7419–24.








