Sigma54-dependent transcription
Yusdi Santoso, Kostas Lymperopoulos, and Achillefs Kapanidis
The objective of this project is to study bacterial σ54-dependent gene transcription at the single-molecule level. We are interested in complex, multi-step transitions characterized by assembly/disassembly events and conformational changes occurring within complexes of transcription proteins and DNA. This project is carried out in collaboration with the group Prof. Martin Buck (Imperial College London) and with Dr Mike Heilemann (Univ of Bielefeld, Germany).
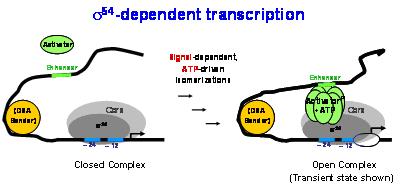
σ54-dependent transcription. Energy from ATP hydrolysis, special DNA binding sequences, and an activator protein are needed for σ54-RNAP to "melt" the DNA around the transcription site and form the RNAP-DNA open complex which can then synthesize RNA.
RNAP-σ54 operates in a different fashion compared to RNAP-σ70, the best studied bacterial RNAP (1). Understanding how RNAP-σ54 works is important from two perspectives. From an engineering perspective, we need to understand how core RNAP is remodelled differently by small proteins (σ70 or σ54) to function in a dramatically different way. From a human-health perspective, σ54-dependent transcription (2) can explain basic elements of human transcription, since the RNAP-σ54 and human transcription machineries are surprisingly similar: to open the DNA helix, both machineries require energy from ATP hydrolysis, specific DNA sequences known as enhancers, and specific activator proteins.
We apply alternating-laser excitation (ALEX) spectroscopy (3), both in solution and on surface-immobilized molecules. Our approach involves the formation of heteroduplex DNA constructs. Base mismatch at various positions in heteroduplex DNA forms short sequences of “melted” DNA, which facilitate the binding of transcription factors or RNAP core/holoenzyme. Complexes with heteroduplex-DNA are stable at sub-nanomolar concentrations, and can be observed using ALEX spectroscopy.
- Buck, M. et al. J. Bacteriol. 182, 4129-4136 (2000)
- Orphanides, G. and Reinberg, D. Cell 108, 439-451 (2002)
- Kapanidis et al., Acc Chem Res, 38, 523-33 (2005)






