DNA-Processing Machines
Site-specific Recombination
Ling Chin Hwang, Nigel D.F. Grindley and Achillefs Kapanidis
Gamma-delta resolvase and Bxb1 integrase are two examples of the serine recombinase family of site specific recombination enzymes–molecular machines that, in a variety of different biological contexts, bring together two DNA sites, break, exchange, and rejoin them to form recombinant sites (1). Recombination occurs within a core synaptic complex that contains two parental DNA sites held together by a tetramer of the recombinase. Formation of this complex, which is generally a tightly regulated process, activates the catalytic activity of the recombinase subunits. Recombination is initiated by catalysis of a double strand break within each parental DNA site–a process that is accompanied by the transient covalent attachment of each recombinase subunit to the 5' end of the broken DNA via a phosphoseryl linkage. Cleavage is followed by DNA strand exchange–a process that appears to involve 180º rotation of one half of the tetramer relative to the other. Recombination is completed by rejoining the DNA ends (now in the recombinant arrangement) by a reversal of the cleavage step; this releases the covalent recombinase-DNA linkage and allows dissociation of the synapse. Our understanding of the recombination process has been substantially illuminated by the solving of several crystal structures of gamma-delta resolvase, most notably including a synaptic complex with a resolvase tetramer and cleaved recombination sites (2). This structure has suggested a molecular mechanism for the strand exchange step. Formation of the synaptic complex creates a novel planar (and hydrophobic) interface that divides the two halves of the synapse, and it is proposed that this interface allows the free rotation of one half relative to the other, while keeping the two halves in close molecular contact.
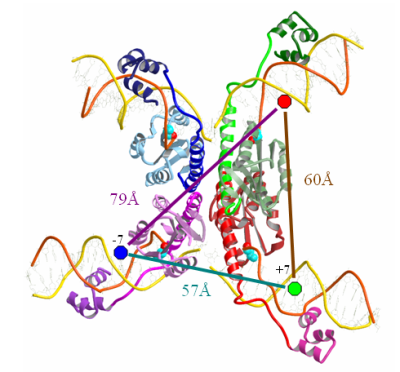
Structure of the gamma-delta resolvase synaptic tetramer complexed with cleaved crossover sites. View through the complex showing the flat interface that separates the complex into two halves. It is proposed that this (hydrophobic) interface allows the free rotation of the half-sites leading to DNA strand exchange. Blue, green and red circles represent fluorophore labels on the DNA strands and their distances between them, as measured from the crystal structure.
The dynamic nature of the recombination process and our inability to synchronize the steps of DNA cleavage, strand exchange and religation provide a challenge to the study of the overall recombination process. To bypass this problem, we have turned to single molecule FRET and alternating laser excitation (ALEX) (3) to identify the intermediates formed during the process and to study the dynamics of their interconversion. For these studies we are using both gamma-delta resolvase and Bxb1 integrase as our model sysems. The advantage of resolvase is that it is the source of all relevant crystal structures (and thus accurate distance measurements). However, the study of resolvase is associated with certain disadvantages. The integrase of phage Bxb1 has several useful attributes: (i) it recognizes two distinct recombination sites, attP and attB, and one of each is required to form a synaptic complex (4). Moreover, the recombination process is unidirectional and irreversible; the recombinant sites, attL and attR, are not effective substrates for the integrase unless an additional protein is present (5). Using substrates either in singly-labeled pairs, or a doubly-labeled site in combination with an unlabeled site, we can detect parental or recombinant sites with characteristic FRET efficiencies and also intermediates that contain cleaved DNA but are held together by the recombinase tetramer. By using recombination sites with mismatched crossover sites, we can trap cleaved intermediates in a form that can never rejoin, but should cycle between the parental and recombinant configurations, exploring all the states in between. We hope to determine the time scale of this cycling process, and from the FRET efficiencies encountered and the lifetimes of any specific FRET states, obtain a picture of the dynamics of the strand exchange process.
This project is carried out in collaboration with Prof. Nigel D.F. Grindley at Yale University.
References:
- Grindley, N.D.F. Whiteson, K.L. and Rice, P.A. (2006) Mechanisms of site-specific recombination. Ann. Rev. Biochem. 75, 567-605.
- Li, W., Kamtekar, S., Xiong, Y., Sarkis, G.J., Grindley, N.D.F. and Steitz, T.A. (2005) Structure of a synaptic gamma-delta resolvase tetramer covalently linked to two cleaved DNAs. Science 309, 1210-1215.
- Lee NK, Kapanidis AN, Wang Y, Michalet X, Mukhopadhyay J, Ebright RH, Weiss S. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys J, 2005, 88, 2939-53.
- Ghosh, P., Pannunzio, N.R. and Hatfull, G.F. (2005) Synapsis in phage Bxb1 integration: selection mechanism for the correct pair of recombination sites. J. Mol. Biol. 349, 331-348
- Ghosh, P. Wasil, L.R. and Hatfull, G.F. (2006) Control of phage Bxb1 excision by a novel recombination directionality factor. PLoS Biol. 4, 964-974






