DNA Polymerase
David Bauer, Cathy Joyce, Johannes Hohlbein, Nigel D.F. Grindley, and Achilles Kapanidis
DNA polymerases are dynamic molecular machines that faithfully copy DNA during DNA replication and repair.[1] One of the important characteristic of this enzyme is its remarkable fidelity, due to a series of noncovalent transitions that precede the chemical step of phosphoryl transfer and serve as kinetic checkpoints, rejecting inappropriate substrates early in the reaction pathway.[2] An important conformational change is the “fingers-closing” transition, inferred from cocrystal structures, in which the addition of the correct complementary deoxyribonucleotide (dNTP) to a polymerase-DNA (Pol-DNA) binary complex results in a transition from an open to a closed conformation, forming a snug binding pocket around the nascent base pair.
The small magnitude of such conformational changes, coupled with the transient nature of the intermediates, makes ensemble study the fingers-closing transitions especially difficult. To get a dynamic view of the DNA polymerase machinery in action, we used single-molecule FRET (smFRET) techniques, including alternating laser excitation (ALEX) and Fluorescence Correlation Spectroscopy (FCS), both in solution as well as in gel.[4, 3]
Single-molecule FRET studies of E. coli DNA polymerase I by Santoso et al.[3] have captured conformational transitions which are crucial for ensuring polymerase accuracy. Using DNA polymerase I (Klenow fragment) labelled with both donor and acceptor fluorophores, we have employed smFRET to study the polymerase conformational transitions that precede nucleotide addition. Our experiments clearly distinguish the open and closed conformations that predominate in Pol-DNA and Pol-DNA-dNTP complexes, respectively. By contrast, the unliganded polymerase shows a broad distribution of FRET values, indicating a high degree of conformational flexibility in the protein in the absence of its substrates; such flexibility was not anticipated on the basis of the available crystallographic structures. Real-time observation of conformational dynamics showed that most of the unliganded polymerase molecules sample the open and closed conformations in the millisecond timescale. Ternary complexes formed in the presence of mismatched dNTPs or complementary ribonucleotides show novel FRET species, which we suggest are relevant to kinetic checkpoints that discriminate against these incorrect substrates.
We are currently expanding our studies to address the mechanisms and kinetics of binding and dissociation of the Pol molecules. This is accomplished by looking at the fluorescence signal due to the binding of freely diffusing Pol (labelled with a donor fluorophore) molecules to surface-immobilised DNA substrate (labelled with an acceptor fluorophore). In particular, we use total internal reflection fluorescence microscopy (TIRF) in combination with ALEX to study hundreds of molecules at the same time for an extended period of time (from several minutes up to half an hour).
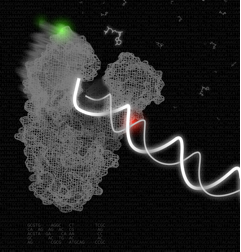
An artistic rendition of the DNA polymerase in action. In this picture, the polymerase-DNA binary complex undergoes the ‘fingers-closing’ transition in the presence of incoming complementary nucleotide. The polymerase-DNA structure shown is based on the B. stearothermophilus DNA polymerase (PDB file 1L3U); the green and red spheres show the positions of the fluorophores.
References
- C.M. Joyce. Encyclopedia of Biological Chemistry, chapter DNA polymerase I, bacterial, pages 720–725. Elsevier, 2004.
- C.M. Joyce and S.J. Benkovic. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry, 43(45):14317–14324, Nov 2004.
- Y. Santoso, C.M. Joyce, O. Potapova, L. Le Reste, J. Hohlbein, J.P. Torella, N.D.F. Grindley, and A.N. Kapanidis. Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc. Natl. Acad. Sci. U. S. A., 107(2):715–720, 2010.
- Y. Santoso and A.N. Kapanidis. Probing Biomolecular Structures and Dynamics of Single Molecules Using In-Gel Alternating-Laser Excitation. Anal. Chem., 81(23):9561–9570, 2009.








